According to two recent research articles published in Nature Genetics, newly created artificial intelligence (AI) systems were able to anticipate the three-dimensional (3D) structure of DNA as well as the function of its regulatory elements based only on their raw sequence.
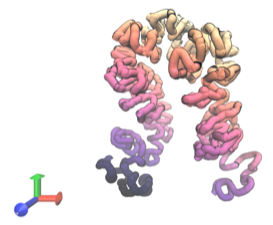 Predicted 3D structure for a segment of human genomic DNA. Image Credit: UT Southwestern Medical Center.
Predicted 3D structure for a segment of human genomic DNA. Image Credit: UT Southwestern Medical Center.
Study author Jian Zhou, PhD., Assistant Professor in the Lyda Hill Department of Bioinformatics at UTSW, believes that these tools could eventually provide new insight into how genetic mutations cause disease as well as new knowledge of how genetic sequence affects the spatial organization and function of chromosomal DNA in the nucleus.
Dr. Zhou is a fellow of the Harold C. Simmons Comprehensive Cancer Center, a scholar at the Cancer Prevention and Research Institute of Texas (CPRIT), and a Lupe Murchison Foundation Scholar in Medical Research.
Taken together, these two programs provide a more complete picture of how changes in DNA sequence, even in noncoding regions, can have dramatic effects on its spatial organization and function.
Jian Zhou, Assistant Professor, Lyda Hill Department of Bioinformatics, UT Southwestern Medical Center
Instructions for building proteins are only encoded in approximately 1% of human DNA. Recent studies have revealed that a large portion of the remaining non-coding genetic material contains regulatory components that control the expression of the coding DNA, such as promoters, enhancers, silencers, and insulators.
According to Dr. Zhou, it is unclear how sequencing affects the majority of the functions of the regulatory elements.
Zhou and his colleagues at Princeton University and the Flatiron Institute created the Sei deep learning model to better comprehend these regulatory elements. Sei correctly classifies these noncoding DNA snippets into 40 “sequence classes” or jobs, such as an enhancer for stem cell or brain cell gene activity.
More than 97% of the human genome is represented by these 40 sequence classes, which were created from approximately 22,000 data sets from earlier research studying genome control. Additionally, Sei can rank each sequence according to its expected activity in each of the 40 types of sequences and forecast how mutations would affect such activities.
The researchers were able to define the regulatory architecture of 47 characteristics and disorders listed in the UK Biobank database and explain how mutations in regulatory components induce certain pathologies by using Sei to human genetics data.
Such abilities can aid in the systematic study of the relationships between changes in genomic sequence and diseases and other features. The results were published in August 2022.
In May 2022, Dr. Zhou announced the creation of a separate tool named Orca, which uses DNA sequences to predict the 3D layout of chromosomes.
Dr. Zhou trained the model to build connections and assessed the model’s capability to predict structure at different length scales using existing datasets of DNA sequences and structural data acquired from prior research that showed the molecule’s folds, twists, and turns.
The results revealed that Orca accurately predicted both small and large DNA structures based on their sequences, particularly for sequences bearing mutations linked to a variety of medical disorders, including a form of leukemia and limb deformities.
The researchers’ use of Orca also allowed them to come up with fresh theories on how the DNA sequence affects both the local and large-scale 3D structure.
Sei and Orca, which are both publicly accessible on web servers and as open-source code, will be used by Dr. Zhou and his team to further investigate the role of genetic mutations in provoking the molecular and physical manifestations of diseases. This research might one day result in new treatment options for these conditions.
The National Institutes of Health (DP2GM146336), CPRIT (RR190071), and the UT Southwestern Endowed Scholars Program in Medical Science provided support for the Orca study.
Journal References:
Chen, K. M., et al. (2022) A sequence-based global map of regulatory activity for deciphering human genetics. Nature Genetics. doi:10.1038/s41588-022-01102-2.
Zhou, J. (2022) Sequence-based modeling of three-dimensional genome architecture from kilobase to chromosome scale. Nature Genetics. doi:10.1038/s41588-022-01065-4.