A study, appearing in the journal Nature Communications, reveals a new system for finding cellular signatures of disease that incorporates robotic platforms for analyzing patient cells with artificial intelligence (AI) approaches for image analysis.
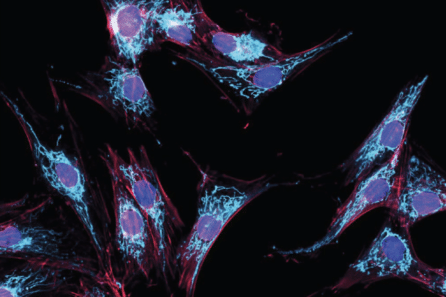 Fibroblasts (cells found in connective tissue) generated by the Array and used to study PD. Image Credit: New York Stem Cell Foundation.
Fibroblasts (cells found in connective tissue) generated by the Array and used to study PD. Image Credit: New York Stem Cell Foundation.
Using their automated cell culture system, researchers at the NYSCF Research Institute worked together with Google Research to find new cellular signatures of Parkinson’s disease by developing and describing over a million skin cell images from a group of 91 healthy controls and patients.
Traditional drug discovery isn’t working very well, particularly for complex diseases like Parkinson’s. The robotic technology NYSCF has built allows us to generate vast amounts of data from large populations of patients, and discover new signatures of disease as an entirely new basis for discovering drugs that actually work.
Susan L. Solomon, JD., CEO, New York Stem Cell Foundation
“This is an ideal demonstration of the power of artificial intelligence for disease research,” added Marc Berndl, Software Engineer at Google Research. “We have had a very productive collaboration with NYSCF, especially because their advanced robotic systems create reproducible data that can yield reliable insights.”
Coupling Artificial Intelligence and Automation
The study made use of NYSCF’s massive repository of patient cells and advanced robotic platform—The NYSCF Global Stem Cell Array® — to describe images of millions of cells from 91 healthy controls and patients with Parkinson’s.
With the Array®, researchers separated and expanded skin cells known as fibroblasts from skin punch biopsy samples, labeled various parts of these cells with a method known as Cell Painting, and developed numerous high-content optical microscopy images.
The subsequent images were inputted into an impartial, AI-powered image analysis pipeline, that detected image characteristics specific to patient cells that could be employed to differentiate them from healthy controls.
These artificial intelligence methods can determine what patient cells have in common that might not be otherwise observable. What’s also important is that the algorithms are unbiased—they do not rely on any prior knowledge or preconceptions about Parkinson’s disease, so we can discover entirely new signatures of disease.
Samuel J. Yang, Research Scientist, Google Research
The need for new hallmarks of Parkinson’s is highlighted by the failure rates in the latest clinical trials for drugs discovered based on precise disease targets and pathways thought to cause the progression of the disease.
The finding of these unique disease signatures using impartial approaches, particularly spanning patient populations, has significance for drug discovery and diagnostics, even illuminating new differences between patients.
“Excitingly, we were able to distinguish between images of patient cells and healthy controls, and between different subtypes of the disease,” noted Bjarki Johannesson, Ph.D., an NYSCF Senior Investigator on the study. “We could even predict fairly accurately which donor a sample of cells came from.”
Applications to Drug Discovery
The Parkinson’s disease hallmarks detected by the researchers can presently be used as a foundation for carrying out drug screens on patient cells, to find out which drugs can reverse these characteristics. The study also offers the largest acknowledged Cell Painting dataset (48TB) as a community resource and can be accessed by the research community (https://nyscf.org/nyscf-adpd/).
In particular, the system is disease-agnostic, only necessitating straightforwardly accessible skin cells from patients. It can also be used for other cell types, such as byproducts of induced pluripotent stem cells that NYSCF develops to model a range of diseases.
The scientists are thus hopeful that their system can open new therapeutic paths for numerous diseases where conventional drug discovery has been ineffective.
This is the first tool to successfully identify disease features with this much precision and sensitivity. Its power for identifying patient subgroups has important implications for precision medicine and drug development across many intractable diseases.
Daniel Paull, PhD., Senior Vice President of Discovery and Platform Development, New York Stem Cell Foundation Research Institute
Journal Reference:
Schiff, L., et al. (2022) Integrating deep learning and unbiased automated high-content screening to identify complex disease signatures in human fibroblasts. Nature Communications. doi.org/10.1038/s41467-022-28423-4.