Jun 20 2019
A new technique has been developed by MIT researchers to obtain more information from images used for training machine-learning models, such as those that can examine medical scans to help identify and treat brain disorders.
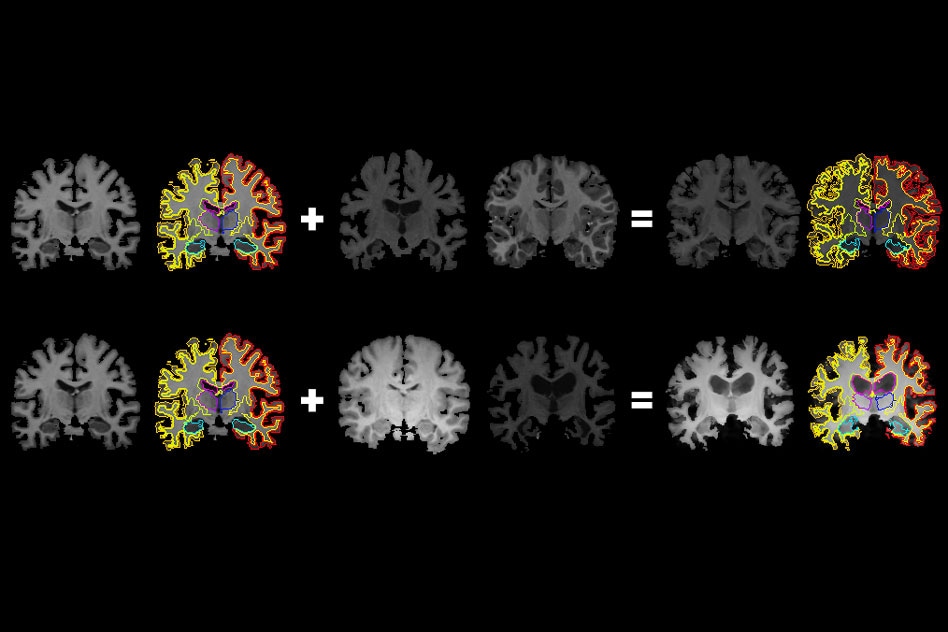 MIT researchers have developed a system that gleans far more labeled training data from unlabeled data, which could help machine-learning models better detect structural patterns in brain scans associated with neurological diseases. The system learns structural and appearance variations in unlabeled scans, and uses that information to shape and mold one labeled scan into thousands of new, distinct labeled scans. (Image credit: MIT)
MIT researchers have developed a system that gleans far more labeled training data from unlabeled data, which could help machine-learning models better detect structural patterns in brain scans associated with neurological diseases. The system learns structural and appearance variations in unlabeled scans, and uses that information to shape and mold one labeled scan into thousands of new, distinct labeled scans. (Image credit: MIT)
Training deep-learning models to detect structural patterns in brain scans linked with neurological disorders and diseases, like multiple sclerosis and Alzheimer’s disease, is a new, active area in medicine.
However, it is tedious to gather the training data; for example, neurological experts have to manually label or separately outline all anatomical structures in each scan. Moreover, in certain cases, like the rare brain conditions in children, only a handful of scans may be available in the initial place.
The MIT researchers presented a paper at the recent Conference on Computer Vision and Pattern Recognition, describing a system that employs a single labeled scan, together with unlabeled scans, to automatically create a large dataset of clear training examples.
With the help of the dataset, machine-learning models can be better trained to locate anatomical structures in the latest scans—more training data will translate to more improved predictions.
The core of the study is to automatically create data for the “image segmentation” process. This process separates an image into regions of pixels that are easier and more meaningful to examine.
The system utilizes a convolutional neural network, or CNN, to achieve this. The network is a machine-learning model that has turned out to be a powerhouse for tasks related to image processing. It examines plenty of unlabeled scans from different equipment and different patients to “learn” contrast, brightness, and anatomical variations.
The network then synthesizes new scans by applying an arbitrary combination of those learned variations to one labeled scan. The newly generated scans are precisely labeled and realistic which are subsequently fed into a varied CNN that learns how to separate new images.
We’re hoping this will make image segmentation more accessible in realistic situations where you don’t have a lot of training data. In our approach, you can learn to mimic the variations in unlabeled scans to intelligently synthesize a large dataset to train your network.
Amy Zhao, Study First Author and Graduate Student, Department of Electrical Engineering and Computer Science and Computer Science and Artificial Intelligence Laboratory, MIT
Zhao further added that the system has attracted some amount of interest, for example, it can be used for training predictive-analytics models at Massachusetts General Hospital, where just one or two labeled scans may be available of predominantly common brain disorders among pediatric patients.
Guha Balakrishnan, a postdoc in CSAIL and EECS; senior author Adrian Dalca, who is also a faculty member in radiology at Harvard Medical School; and EECS professors John Guttag and Fredo Durand, also contributed to the paper.
The “Magic” behind the system
While the system is currently used in medical imaging, it was originally started as a method to generate training data for a smartphone app that can possibly detect and retrieve data about cards from the famous collectable card game called “Magic: The Gathering.” This game was launched in the early 1990s and has over 20,000 special cards, with more numbers of them released every few months. These cards can be used by players for building custom playing decks.
A keen “Magic” player, Zhao intended to create a CNN-powered app that captured an image of any card using a smartphone camera and then automatically extracted data such as rating and price from online card databases.
“When I was picking out cards from a game store, I got tired of entering all their names into my phone and looking up ratings and combos,” Zhao stated. “Wouldn’t it be awesome if I could scan them with my phone and pull up that information?”
However, she realized that's an extremely difficult computer-vision training task.
You’d need many photos of all 20,000 cards, under all different lighting conditions and angles. No one is going to collect that dataset.
Amy Zhao, Study First Author and Graduate Student, Department of Electrical Engineering and Computer Science and Computer Science and Artificial Intelligence Laboratory, MIT
To make the CNN learn on how to warp a card into different positions, Zhao trained the network on a smaller dataset of about 200 cards, with 10 clear photos of each card. The network calculated different reflections, angles, and lighting—for when cards are located in plastic sleeves—to the synthesized realistic warped forms of any card in the dataset. It was truly a fascinating project, “but we realized this approach was really well-suited for medical images, because this type of warping fits really well with MRIs,” Zhao stated.
Mind warp
Magnetic resonance images, or MRIs, in short, are made up of 3D pixels, known as voxels. When experts are separating MRIs, they isolate and label voxel areas on the basis of the anatomical structure containing them.
The scans’ diversity, induced by the differences in each brain and the equipment utilized, makes it difficult to apply machine learning to automate this procedure.
There are some existing techniques that use “data augmentation” to synthesize training examples from labeled scans. Data augmentation warps labeled voxels into varied positions. However, such techniques need experts to manually write numerous augmentation guidelines, and certain synthesized scans do not resemble a real human brain, which may be unfavorable to the learning process.
Instead, the system developed by the researchers automatically learns how to create realistic scans. The system was also trained on 100 unlabeled scans from actual patients to calculate spatial changes—anatomical correspondences from one scan to another.
This results in many numbers of “flow fields,” which model the way voxels pass from scan to scan. At the same time, it calculates intensity changes, which capture appearance differences induced by various factors, including noise and image contrast.
While synthesizing a new scan, the system applies an arbitrary flow field to the original labeled scan, which moves around voxels until it structurally corresponds to an actual, unlabeled scan.it then THEN overlays a haphazard intensity transformation. Then, the system finally maps the labels to the new structures, by tracking the way the voxels shift in the flow field. Towards the end, the generated scans closely look like the true, unlabeled scans, but with precise labels.
In order to test their automated segmentation precision, the investigators utilized Dice scores, which determine the way a single 3D shape accurately fits over another, on a scale of 0 to 1. The researchers compared their system to conventional segmentation techniques—both automated and manual—on 30 diverse brain structures across a total of 100 held-out test scans. Huge structures were found to be comparably precise among all the techniques. Yet, the researchers’ system surpassed all other techniques on smaller structures, for example, the hippocampus, which takes up only around 0.6% of a brain, in terms of volume.
That shows that our method improves over other methods, especially as you get into the smaller structures, which can be very important in understanding disease. And we did that while only needing a single hand-labeled scan.
Amy Zhao, Study First Author and Graduate Student, Department of Electrical Engineering and Computer Science and Computer Science and Artificial Intelligence Laboratory, MIT
Following the approval of the study’s “Magic” roots, the code which is named after one of the game’s cards, “Brainstorm,” is publicly available on Github