Jun 7 2019
About half a million children around the world are born with a rare hereditary disease each year. Getting a definitive diagnosis can be hard and time consuming. In a study involving 679 patients having 105 different rare diseases, researchers from the University of Bonn and the Charité - Universitätsmedizin Berlin have demonstrated that artificial intelligence (AI) can be applied to diagnose rare diseases more reliably and efficiently. A neural network automatically integrates portrait photos with genetic and patient data. The results are currently published in the journal “Genetics in Medicine.”
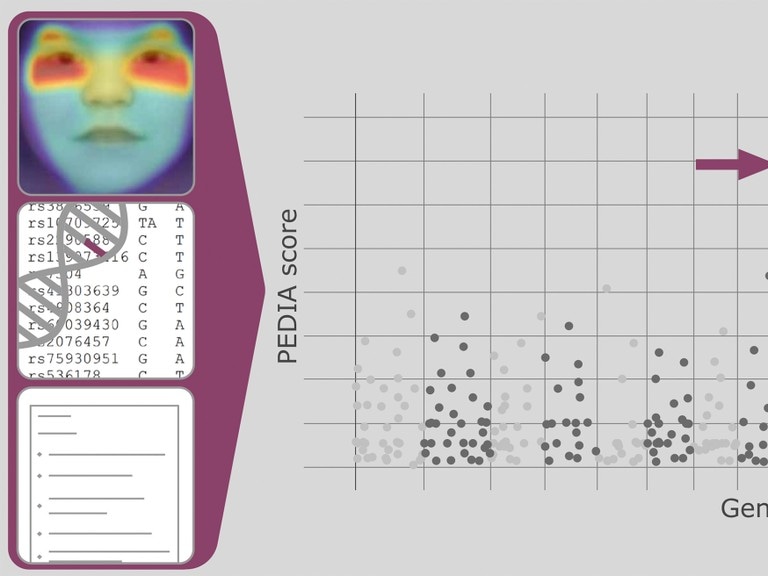 With artificial intelligence to a diagnosis of rare hereditary diseases: The neural network combines data from portrait images with gene and patient data. (© Photo: Tori Pantel)
With artificial intelligence to a diagnosis of rare hereditary diseases: The neural network combines data from portrait images with gene and patient data. (© Photo: Tori Pantel)
A number of patients with rare diseases experience prolonged trials and tribulations until they are properly diagnosed. "This results in a loss of valuable time that is actually needed for early therapy in order to avert progressive damage," explains Prof. Dr. med. Dipl. Phys. Peter Krawitz from the Institute for Genomic Statistics and Bioinformatics at the University Hospital Bonn (UKB). Along with an international team of researchers, he shows how AI can be applied to make comparatively fast and dependable diagnoses in facial analysis.
The scientists used data of 679 patients with 105 different diseases caused by the variation in a single gene. These comprise of, for example, mucopolysaccharidosis (MPS), which causes stunted growth, bone deformation, and learning difficulties. Mabry syndrome also leads to intellectual disability. All these diseases have in common that the facial features of those afflicted exhibit irregularities. This is mainly characteristic, for example, of Kabuki syndrome, which is suggestive of the make-up of a traditional Japanese form of theatre. The eyebrows are arched, the spaces between the eyelids are long, and the eye-distance is wide.
The used software can automatically spot these typical features from a photo. Along with the clinical symptoms of the patients and genetic data, it is possible to compute with high accuracy which disease is most likely to be the cause. The AI and digital health company FDNA has created the neural network DeepGestalt, which the scientists use as an AI tool for their work.
PEDIA is a unique example of next-generation phenotyping technologies. Integrating an advanced AI and facial analysis framework such as DeepGestalt into the variant analysis workflow will result in a new paradigm for superior genetic testing.
Dekel Gelbman, CEO, FDNA
Researchers train the neural network with 30,000 images
The researchers trained this computer program with about 30,000 portrait pictures of people having rare syndromal illnesses. "In combination with facial analysis, it is possible to filter out the decisive genetic factors and prioritize genes," says Krawitz. "Merging data in the neuronal network reduces data analysis time and leads to a higher rate of diagnosis.”
The head of the Institute of Genomic Statistics and Bioinformatics at the UKB has been partnering with FDNA for quite a while. "This is of great scientific interest to us and also enables us to find a cause in some unsolved cases," said Krawitz. Many patients are presently still seeking a reason for their symptoms.
The research is a team work between computer science and medicine. This can also be observed in the shared first authorship of the computer scientist Tzung-Chien Hsieh, doctoral student at the institute of Professor Krawitz, and Dr. Martin Atta Mensah, physician at the Institute of Medical Genetics and Human Genetics of the Charité and Fellow of the Clinician Scientist Program of the Charité and Berlin Institute of Health (BIH). Prof. Dr. Stefan Mundlos, Director of the Institute of Medical Genetics and Human Genetics at the Charité, also took part in the research, as did more than 90 other researchers.
Patients want a prompt and accurate diagnosis. Artificial intelligence supports physicians and scientists in shortening the journey. This also improves the quality of life of those affected to some extent.
Dr. Christine Mundlos, Deputy Managing Director, Alliance of Patients with Chronic Rare Diseases (ACHSE) e.V.
Results will be presented at international conference
At the conference of the European Society of Human Genetics (ESHG) held between 15th and 18th June in Gothenburg (Sweden), the researchers will present their study. FDNA will also be at the conference.