Jun 21 2018
Medical image registration is a typical method where two images, such as magnetic resonance imaging (MRI) scans are overlaid in order to compare and examine anatomical variances in depth. If a patient has a brain tumor, for example, the physician can overlap a brain scan from earlier months onto the latest scan to examine small variations in the tumor’s progress.
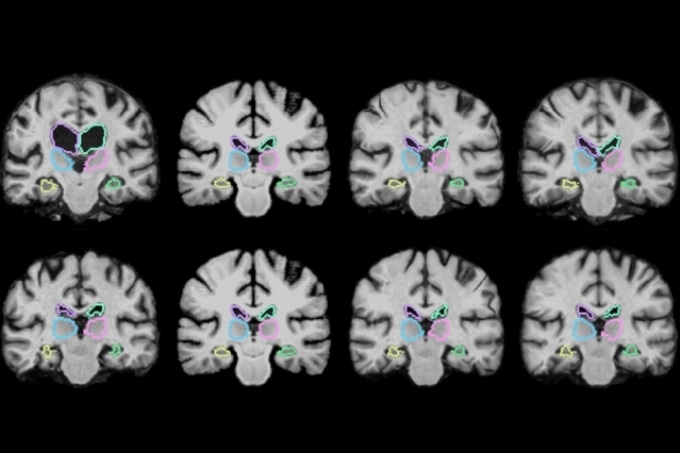 MIT researchers describe a machine-learning algorithm that can register brain scans and other 3D images more than 1,000 times more quickly using novel learning techniques. (Courtesy of the researchers)
MIT researchers describe a machine-learning algorithm that can register brain scans and other 3D images more than 1,000 times more quickly using novel learning techniques. (Courtesy of the researchers)
This process, however, can mostly take two hours or more, as traditional systems precisely align each of potentially a million pixels in the combined scans. In two upcoming conference papers, MIT scientists illustrate a machine-learning algorithm that can register brain scans and other 3D images over 1,000 times more rapidly using novel learning methods.
The algorithm functions by “learning” while registering numerous pairs of images. In doing so, it obtains information about how to align images and estimates some ideal alignment parameters. After training, it applies those parameters to map all pixels of a single image to another, all at once. This decreases registration time to a minute or two using a regular computer, or less than a second using a GPU with similar accuracy to advanced systems.
“The tasks of aligning a brain MRI shouldn’t be that different when you’re aligning one pair of brain MRIs or another,” says co-author on both papers Guha Balakrishnan, a graduate student in MIT’s Computer Science and Artificial Intelligence Laboratory (CSAIL) and Department of Engineering and Computer Science (EECS). “There is information you should be able to carry over in how you do the alignment. If you’re able to learn something from previous image registration, you can do a new task much faster and with the same accuracy.”
The papers will be presented at the Conference on Computer Vision and Pattern Recognition (CVPR), held this week, and at the Medical Image Computing and Computer Assisted Interventions Conference (MICCAI), held in September 2018. Co-authors are Adrian Dalca, a postdoc at Massachusetts General Hospital and CSAIL; Amy Zhao, a graduate student in CSAIL; Mert R. Sabuncu, a former CSAIL postdoc and now a professor at Cornell University; and John Guttag, the Dugald C. Jackson Professor in Electrical Engineering at MIT.
Retaining information
MRI scans are essentially numerous stacked 2D images that form huge 3D images, known as “volumes,” comprising a million or more 3D pixels, known as “voxels.” Thus, it’s highly time-consuming to align all voxels in the first volume with the voxels in the second. Furthermore, scans can come from various machines and have varied spatial orientations, meaning matching voxels are even more computationally multifaceted.
You have two different images of two different brains, put them on top of each other, and you start wiggling one until one fits the other. Mathematically, this optimization procedure takes a long time.
Adrian Dalca, Senior Author on the CVPR paper and the MICCAI paper’s lead author
This process becomes predominantly slow when examining scans from large populations. Neuroscientists examining differences in brain structures spanning numerous patients with a specific disease or condition, for example, could possibly require hundreds of hours.
That’s because those algorithms have one key error: They do not learn. After each registration, they terminate all data relating to voxel location. “Essentially, they start from scratch given a new pair of images,” Balakrishnan says. “After 100 registrations, you should have learned something from the alignment. That’s what we leverage.”
The scientists’ algorithm known as “VoxelMorph” works using a convolutional neural network (CNN), which is a machine-learning approach typically used for image processing. These networks comprise a number of nodes that process image and other information across a number of layers of computation.
In the CVPR paper, the scientists trained their algorithm on 7,000 publicly available MRI brain scans and then analyzed it on 250 extra scans.
In the training phase, brain scans were loaded into the algorithm in pairs. Using a CNN and adapted computation layer known as a spatial transformer, the technique captures similarities of voxels in a single MRI scan with voxels in the other scan. In doing so, the algorithm learns information relating to groups of voxels—such as anatomical shapes typical to both scans—which it uses to calculate enhanced parameters that can be applied to any scan pair.
When fed two new scans, a basic mathematical “function” uses those enhanced parameters to quickly calculate the precise alignment of every voxel in both scans. In a nutshell, the algorithm’s CNN component gains all essential information during training so that, during each new registration, the whole registration can be achieved using one, easily computable function evaluation.
The scientists found their algorithm could exactly register all of their 250 test brain scans—those registered after the training set - in just two minutes using a traditional central processing unit, and within one second using a graphics processing unit.
Significantly, the algorithm is “unsupervised,” meaning it does not require extra information beyond image data. Some registration algorithms integrate CNN models but require a “ground truth,” meaning another traditional algorithm is initially run to compute exact registrations. The scientists’ algorithm maintains its correctness without that data.
The MICCAI paper develops an improved VoxelMorph algorithm that “says how sure we are about each registration,” Balakrishnan says. It also ensures the registration “smoothness,” meaning it does not create holes, folds, or basic distortions in the composite image. The paper showcases a mathematical model that confirms the algorithm’s accuracy using something known as a Dice score, a typical metric to assess the accuracy of overlapped images. Across 17 brain regions, the improved VoxelMorph algorithm scored the same accuracy as a normally used modern registration algorithm, while providing methodological and runtime improvements.
Beyond brain scans
Besides examining brain scans, the speedy algorithm has a broad range of possible applications, the scientists say. MIT colleagues, for example, are presently using the algorithm on lung images.
The algorithm could also open doors to enable image registration during operations. Various scans of diverse speeds and qualities are presently used before or during some surgeries. But those images are not registered until post-operation. When resecting a brain tumor, for example, surgeons occasionally scan a patient’s brain before and after surgery to see if they have removed the entire tumor. If any bit is still present, they return to the operating room.
With the new algorithm, Dalca says, surgeons could possibly register scans in near real-time, acquiring a much richer picture on their progress. “Today, they can’t really overlap the images during surgery, because it will take two hours, and the surgery is ongoing” he says. “However, if it only takes a second, you can imagine that it could be feasible.”