Jul 15 2016
A team of scientists has developed a model that can predict bat species most likely to transmit Ebola and other filoviruses. Findings highlight new potential hosts and geographic hotspots worthy of surveillance. So reports a new paper in the journal PLoS Neglected Tropical Diseases.
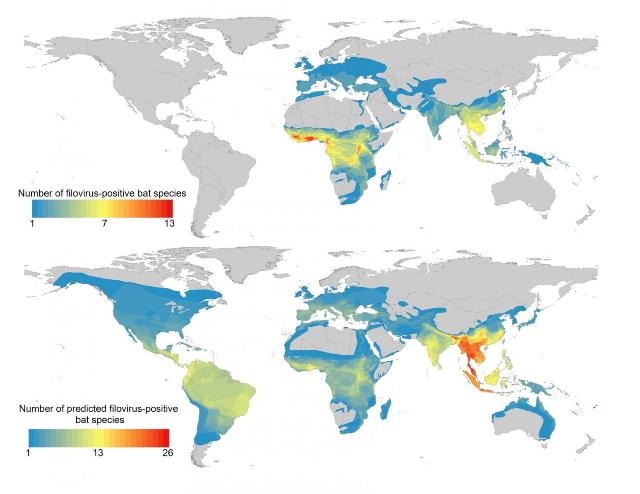 This is a map of known and predicted bat hosts of filoviruses, showing hotspots in Southeast Asia. (CREDIT: PLOS Neglected Tropical Diseases by Han et al. from the Cary Institute of Ecosystem Studies)
This is a map of known and predicted bat hosts of filoviruses, showing hotspots in Southeast Asia. (CREDIT: PLOS Neglected Tropical Diseases by Han et al. from the Cary Institute of Ecosystem Studies)
Filoviruses have devastating effects on people and primates, as evidenced by the 2014 Ebola outbreak in West Africa. For nearly 40 years, preventing spillover events has been hampered by an inability to pinpoint which wildlife species harbor and spread the viruses. To date, outbreaks have occurred in areas with exceptionally high biodiversity.
Barbara Han, a disease ecologist at the Cary Institute of Ecosystem Studies and the paper's lead author, comments, "Using machine learning methods developed for artificial intelligence, we were able to bring together data from ecology, biogeography, and public health to identify bat species with a high probability of harboring Ebola and other filoviruses. Understanding which species carry these viruses, and where they are located, is essential to preventing future spillovers."
The research team included scientists from the University of Georgia, Massey University, and the University of California. They focused on bats - primary suspects in the search for Ebola reservoirs. While Ebola virus has never been isolated from a live African mammal in the wild, several bat species have tested positive for filovirus antibodies. In lab inoculations, three bat species have replicated Ebola. And, unlike great apes and humans, bats infected with filoviruses are largely symptom-free.
First, the team developed a 'profile' of filovirus-positive bat species by looking at life history, physiological, and ecological attributes of the 21 bat species known to harbor filoviruses. Using 57 variables, from diet and reproductive behavior to migratory patterns and species density, an algorithm learned features that distinguish bats that have tested positive for filoviruses from other bat species with 87% accuracy.
David Hayman of Massey University notes, "The model allows us to move beyond our own biases and find patterns in the data that only a machine can. Instead of predicting where Ebola and other filovirus outbreaks will occur by looking at the last spillover event, it forecasts risk based on the intrinsic traits of filovirus-positive bat species."
Those traits include: early maturity, having more than one pup per year (most bats only have one), offspring that are large at birth, and a tendency to live in large groups. Compared to other bats, filovirus-positive species also have broader geographic ranges that overlap with a higher diversity of mammal species per square kilometer.
When data on the world's 1116 bat species were searched using this filovirus-positive bat profile, machine learning identified new potential hosts based on their traits. Once mapped, these bats were more widely distributed than the team expected. While many potential bat hosts are found in sub-Saharan Africa, they also range across Southeast Asia and Central and South America.
Han explains, "Our results corroborate studies in Africa that have predicted the environmental niche of Ebola spans the primary tropical rainforest. But in a departure from past research, we identified several hotspots in Southeast Asia where up to 26 potential reservoir species overlap, notably in Thailand, Burma, Malaysia, Vietnam, and northeast India."
John Drake of the University of Georgia concludes, "Maps generated by the algorithm can help guide targeted surveillance and virus discovery projects. We suspect there may be other filoviruses waiting to be found. An outstanding question for future work is to investigate why there are so few filovirus spillover events reported for humans and wildlife in Southeast Asia compared to equatorial Africa."