A developing system that integrates quick imaging with artificial intelligence could assist researchers in constructing an extensive picture of present and historic environmental change—by swiftly and precisely examining pollen.
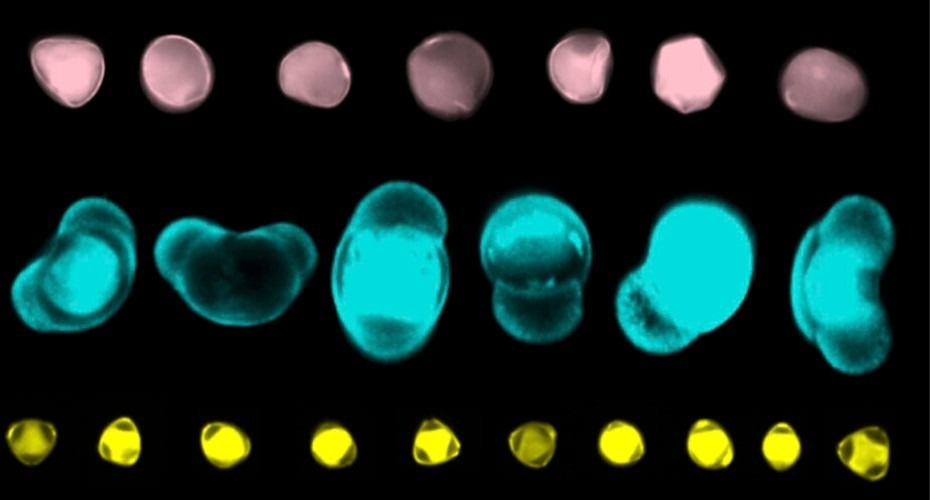
Image Credit: University of Exter.
Pollen grains from various plant species are unique and recognizable depending on their shape. Examining which pollen grains have been captured in samples like sediment cores from lakes helps researchers comprehend which plants were blooming at any given point in history, possibly dating back thousands to millions of years.
So far, researchers have manually counted pollen types in sediments or from air samples utilizing a light microscope—a specialized and tedious task.
Currently, researchers at the University of Exeter and Swansea University are integrating state-of-the-art technologies such as imaging flow cytometry and artificial intelligence to construct a system that has the potential to determine and classify pollen at much quicker rates.
At present, their development is reported in a research study in New Phytologist. As well as building an entire picture of past flora, the research group believes the technology could be applied to highly accurate pollen readings in today’s surroundings, which might aid hayfever sufferers in reducing symptoms.
Pollen is an important environmental indicator, and piecing together the jigsaw of different pollen types in the atmosphere, both today and in the past, can help us build up a picture of biodiversity and climate change. However, recognizing what plant species pollen belongs to under a microscope is incredibly labor-intensive and cannot always be done.
Dr Ann Power, Biosciences, University of Exeter
Power added, “The system we’re developing will cut the time this takes dramatically and improve classifications. This means we can build a richer picture of pollen in the environment far more swiftly, revealing how the climate, human activity, and biodiversity have changed over time, or better understand what allergens are in the air we breathe.”
Earlier, the research group utilized the system to automatically examine a 5,500-year-old slice of lake sediment core, quickly categorizing over a thousand pollen grains. In the past, this would have taken a specialist up to eight hours to count and classify—a task the new system finished in under an hour.
The new system utilizes imaging flow cytometry, a technology normally utilized to examine cells in medical research, to rapidly capture pollen images.
A special kind of artificial intelligence has been developed depending on deep learning to determine the various types of pollen in an environmental sample. This can make such distinctions even when the sample has been imperfect.
Up to now, the AI systems in development to categorize pollen learn from and test on the same pollen libraries – which means each sample is perfect and belongs to species previously seen by the network.
Dr Claire Barnes, Swansea University
Barnes continued, “These systems are not able to recognize pollen from the environment that’s taken some knocks along the way, nor to categorize pollen not included in training libraries. Incorporating a unique version of deep learning into our system means the artificial intelligence is smarter and applies a more flexible approach to learning.”
“It can deal with poor quality images and can use shared species characteristics to predict what family of plant the pollen belongs to even if the system hasn’t seen it before during training,” added Barnes.
In the years to come, the research believes in improving and launching the new system and utilizing it to learn more about grass pollen, a particular irritant for hayfever sufferers.
Some grass pollens are more allergenic than others. If we can understand better which pollens are prevalent at specific times, it would lead to improvements in the pollen forecast that could help people with hayfever plan to reduce their exposure.
Dr Ann Power, Biosciences, University of Exeter
Journal Reference:
Barnes, C. M., et al. (2023) Deductive automated pollen classification in environmental samples via exploratory deep learning and imaging flow cytometry. New Phytologist. doi.org/10.1111/nph.19186.