For studying proteins, a new method has been developed by scientists at Martin Luther University Halle-Wittenberg (MLU) and the European Molecular Biology Laboratory in Hamburg.
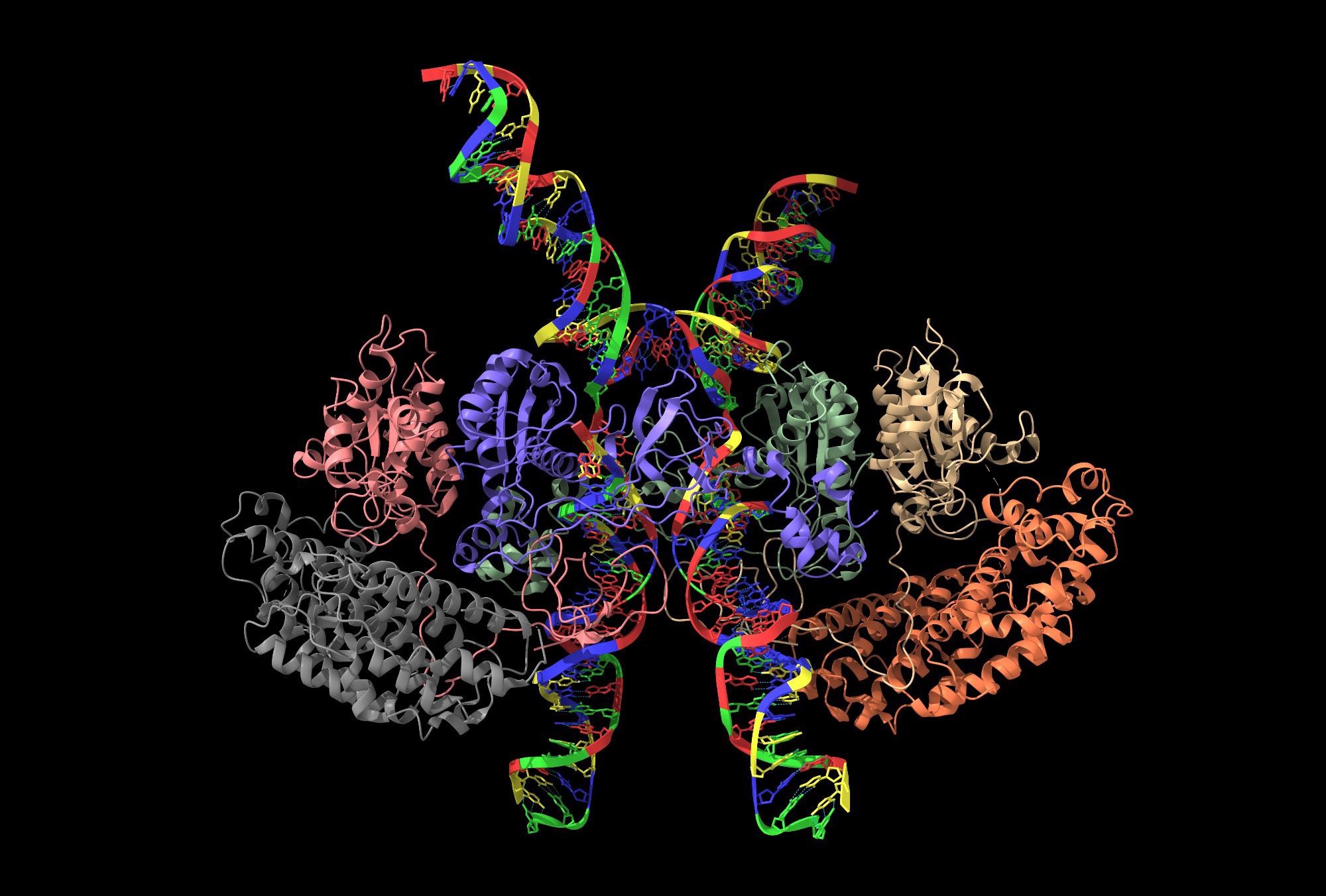
Image Credit: Shutterstock.com/ Volodymyr Dvornyk
The researchers efficiently developed an AI-based approach for analyzing data from cryo-electron microscopy. In the coming days, this will allow several protein complexes to be analyzed directly in cells at the same time. The work was published in the scientific journal Structure.
Cryo-electron microscopy is a fairly new method used to examine the structure of materials, proteins or cells. Theoretically, it works this way: Samples are immediately frozen and bombarded with electrons. Initially, two-dimensional images are created with microscopes, which could be assembled into a 3D model.
For proteins, the microscopy enables highly elaborate images of the structure to be created at the level of single-atom. Understanding the protein structure is vital.
Proteins are the workhorses of the cell; they control all of the important processes—from bone growth to metabolism. However, the role a protein plays and how it functions cannot be precisely understood until its structure is known.
Panagiotis Kastritis, Assistant Professor, Centre for Innovation Competence HALOmem, Martin-Luther-University Halle-Wittenberg
In turn, this knowledge could be utilized in creating therapies for several diseases such as cancer or Alzheimer’s.
But, cryo-electron microscopy offers only three-dimensional structures, and it cannot identify proteins to categorize their identity. The data needs to be interpreted by the scientists themselves, which can lead to possible errors.
The 3D structure of some proteins is nearly identical even though they have completely different functions in the body. When researchers use their experience to decide how to interpret the data and identify the proteins, this can greatly impact the results of their experiments.
Ioannis Skalidis, Doctoral Student, Martin-Luther-University Halle-Wittenberg
Skalidis is also a member of Kastritis’ research group.
Hence, researchers developed a new approach that interprets the raw data from the electron microscope automatically. To perform this, they used many open-source programs, some of which have been developed by themselves, as well as through DeepMind’s AlphaFold.
In July 2021, its developers stated that they could accurately forecast the structure of most human proteins. It was regarded as one of the trickiest puzzles of molecular biology until then.
In the new research, scientists from Halle and Hamburg analyzed protein complexes as they occur in cells, instead of looking at isolated proteins.
Proteins generally do not function on their own, but in larger groups. The structure of their binding sites changes depending on what the proteins are working alongside. That is why it is so important to study them under conditions that are as realistic as possible.
Panagiotis Kastritis, Assistant Professor, Centre for Innovation Competence HALOmem, Martin-Luther-University Halle-Wittenberg
The novel combination of approaches used by the team will also allow other research groups to analyze the structures of such protein complexes more easily in the future.
The study was financially supported by the Federal Ministry of Education and Research, the European Regional Development Fund (ERDF), Saxony-Anhalt, and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation).
Journal Reference:
Skalidis, I., et al. (2022) Cryo-EM and artificial intelligence visualize endogenous protein community members. Structure. doi.org/10.1016/j.str.2022.01.001.